Assays and Services
The HIMC pair efficient study and sample management with a comprehensive suite of immune monitoring assays that allow us to quantify circulating proteins, phenotypically and functionally characterize immune cells at the proteomic and transcriptomic level and map their spatial organization in tissues. HIMC offers the most advanced equipment in the field for research at Genomic, cellular and proteomic analysis. Driven by a combination of deep scientific and technical expertise and innovative technology, all of our assays are developed to balance innovation with robust protocols and operating procedures to ensure data quality and reproducibility.
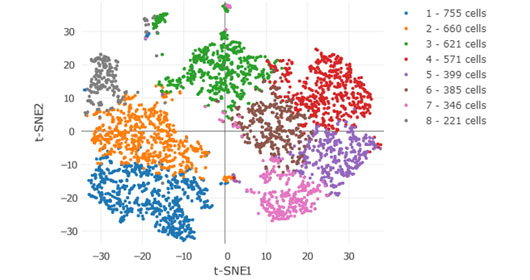
The HIMC computational analysis team leverages Mount Sinai’s High Performance Computing cluster, Minerva, as well as Amazon Web Services (AWS) to perform high throughput data processing and analysis. We have developed a range of efficient semi-automated data analysis pipelines that aim to facilitate analysis and interpretation of complex immunological datasets. For example, we have developed a CyTOF data processing and analysis pipeline that includes automated quality control and single and multi-sample data analysis using state-of-the-art algorithms such as Phenograph for community clustering, viSNE for dimensionality reduction, and association analytics using regularized regression to identify correlates with experimental or clinical features in the data. Our single-cell RNA-seq pipeline includes quality control and the 10X Genomics data processing pipeline. Our computational pipelines can be run in parallel on Minerva or on Amazon Web Services as Dockerized batch jobs, and the data securely can be shared with researchers via cloud-based tools (e.g. AWS, GitHub, Mt. Sinai Box). We are also developing analysis pipelines (e.g. reproducible Jupyter notebooks) for Illumina and Olink proteomics datasets. The HIMC also supports an institutional enterprise Cytobank account; a cloud-based platform that facilitates storage, analysis, visualization and sharing of cytometric data sets.
The HIMC is also actively developing novel interactive web-based data sharing tools such as the CyTOF Data Viewer. The following example illustrates a CyTOF analysis of the immune infiltration in a hepatocellular carcinoma specimen following anti-PD1 therapy. The dataset can be used to interactively browse Phenograph clustering results and viSNE scatterplots to view immune populations and protein expression patterns that are differentially enriched in the tumor, border and adjacent liver tissue.
The HIMC leverages our deep immunological and technical expertise to offer consultation in developing study-specific biospecimen collection protocols and innovative immunologic assessment plans and approaches to evaluate immune responses in clinical studies. We will help define the endpoints of the immune assay, assay parameters, sample requirements (collection, labeling, storage, handling, and shipping) and processing, reagents and controls, data analysis, data management expectations, and estimated costs. We encourage you to contact us as early as possible when designing a study to see how we can help with the process.
Initial Study Consultation
Request Form
Our staff of scientists and technical specialists can offer ongoing expert advice throughout a project to assist investigators in designing and conducting optimal experiments, preparing appropriate samples for assays, and analyzing and interpreting results. We rely on our extensive expertise to perform complex immune assays with a focus on maximizing data quality to reproducibility, and leverage our bulk purchasing power and relationships with vendors for efficiency and cost-effectiveness.
- Blood and Biospecimen Processing
The HIMC offers a full range of biospecimen processing services including processing, isolation and secure storage of serum, plasma, and mononuclear cells isolated from peripheral blood, bone marrow and tissues. We also offer high throughput automated DNA/RNA isolation using a QiaSymphony system.
- Cell Sorting and Enrichment
The HIMC offers targeted magnetic enrichment of viable immune populations using our RoboSep platform for a range of downstream assays, such as nucleic acid isolation, or downstream functional assays. More complex cell sorting experiments can be performed in collaboration with the Flow Cytometry CoRE, which is located directly adjacent to the HIMC and offers a BSL2+ FACS facility equipped with three 5-laser BD FACS Aria cell sorters.
- Sample Management
The HIMC has implemented a HIPAA compliant secure bar-coded sample management and tracking system using Freezerwork (v6.1) supported by the ISMMS secure server. This system ensures the crucial sample data management in a reliable maintaining sample quality for biomedical research, as well as getting the most valuable samples to the researchers in a timely and efficient way such that; ship samples to other sites in a streamlined manner, check out/check in feature manages sample usage and availability and meet all regulatory requirements with an automated audit trail. All the samples are bar-coded in a unique identification number using the bar code printer and placed the dedicated storage locations. All storage freezers (-20 and -80 oC freezers and LN2 tanks) are installed with temperature monitor “Smart Vue” for the centralized live tracking/remote freezer log-in for 24/7/365 security for the temperature fluctuation. Frozen specimens can be shipped to other institutions using dry-shippers equipped with temperature sensors.

- Proseek Multiplex Using Proximity Extension Assay (Olink Platform)
The HIMC has established a platform for highly multiplexed profiling of serum/plasma cytokines leveraging O-Link Proteomics and our Fluidigm Biomark HD microfluidic PCR system allowing robust high-throughput screening for soluble protein biomarkers using very small amounts of sample input. These immunoassays allow measurement of 92 proteins across 96 samples simultaneously using only one microliter of serum, plasma, or almost any other type of biological sample. The Proximity Extension Assay (PEA) technology that underlies these assays makes relies on dual antibody recognition and incorporates several innovative QC steps to evaluate each step of the assay protocol, resulting in an extremely robust assay. Critically, in addition to profiling cytokines, chemokines and growth factors, this platform also allows detection of circulating immune co-stimulatory and inhibitory molecules.
- Multiplex Soluble Factor Analysis Using the Luminex system
Luminex based assays are used to measure biomarkers, cytokines, chemokines, hormones, nitric oxide and growth factor, cell signaling and phospho-protein detection in pathway specific for immune call activation. HIMC is equipped with the high-throughput Luminex INTELLIFLEX and the Luminex 200 system. The xMAP® INTELLIFLEX is a new compact, versatile, and flow-based multiplexing platform, able to detect up to 500 analytes in a single sample volume, compatible for both 96-well and 384-well format. It also has broader dynamic range which reduces the number of out-of-range values. With the proven performance of xMAP technology and enhanced performance, the INTELLIFLEX instrument empowers assay development and simplify the user experience. Our Luminex 200 system is for low throughput multiplexed platform, capable of detect up to 41 analytes simultaneously in a single well of microtiter plate with Data analysis via X-ponent 3.1.
- ELLA Automated and Real-time Cytokine Analysis
HIMC is equipped with two ELLA Automated immunoassay systems. The Ella platform eliminates the tedious setup and lengthy incubations typical of traditional plate-based immunoassays. The automated microfluidic cartridge design minimizes user error while ensuring highly precise and reproducible results across multiple users and sites. Once the immunoassay run is initiated, Ella proceeds from start to finish without the need for manual intervention. Ella pneumatically interfaces with the Simple Plex assay cartridge to perform all reagent additions, wash steps, and incubations under precisely controlled conditions. Similar to a pre-kitted ELISA, all reagents including matched antibody pairs have been validated. But with Ella, the reagents are pre-loaded onto a cartridge for ease of use. No standard curve preparation is needed. Running an assay simply requires adding diluted samples and wash buffer to the cartridge before placing - Grand Serology (ELISA) for Characterized Antigens
Serum or plasma from peripheral blood is used to test for presence of IgG responses to defined antigens. Currently, a tumor panel of defined tumor antigens is available (includes NY-ESO-1, SSX2, Melan-A, TP53 etc.), though the test is easily customizable to include additional antigens of choice, or to test for other subclasses and isotypes such as IgE or IgG4. The assay is an ELISA (enzyme-linked immunosorbent assay) with full-length proteins or overlapping peptides, to determine primarily linear epitopes. The sensitivity of the assay is very high, requiring only microliters of serum or plasma, reproducible, controlled, and results are reported following titration to allow for quantitative comparisons between samples. This flexible assay can also be adapted for ascites, urine, or supernatants of cell cultures where immunoglobulin may be present. - Seromics: Protein Array Antibody Profiling
Seromics is an exploratory, hypothesis-generating platform that allows testing thousands of human proteins simultaneously as potential targets of autoantibodies from patient serum or plasma. The method only requires few microliters of material, and may be customized for applications other than IgG detection. It is ideally suited to comprehensively look for serum antibody changes at the antigen-specific level following treatment, for example immunotherapy, vaccine, chemotherapy, or radiotherapy. Results are normalized and expressed as fold change from pre- to post-treatment. Alternatively, the seromic platform can be used to define biomarkers or sets of antigens present at baseline in specific patient populations, and that could be used as prognostic or predictive markers. This approach requires a minimal number of samples (20) to be tested in order to establish cutoffs of specific responses. Data analysis is provided, and results are expressed as a list of antigens scored for their likelihood of being immunogenic, spontaneously or as a change related to treatment.
- High Dimensional Mass Cytometry (CyTOF)
Mass cytometry merges the high-throughput single cell analysis of flow cytometry with the specificity and resolution of mass spectrometry to allow highly multi-parametric single cell analysis of heterogeneous cell samples. The HIMC leads one of the world’s most active and comprehensive mass cytometry immune monitoring programs, serving multiple investigators and studies at ISMMS as well as other academic and industry collaborations. The center currently houses three Helios mass cytometer. The HIMC has integrated these instruments as part of a comprehensive mass cytometry pipeline supported by three dedicated technical specialists and includes optimized sample processing SOPs, a reagent bank of over 500 metal-conjugated antibodies, a range of optimized immune profiling panels, custom antibody conjugation services and an efficient data processing pipeline to facilitate analysis and integration of complex cytometry datasets. We have extensive experience with all aspects of the mass cytometry workflow, having processed over 5000 samples for mass cytometry studies spanning a diverse range of sample types.
Importantly, mass cytometry can be used for comprehensive phenotypic characterization of single cells in biospecimens, but it can also be used for sophisticated functional studies such as multiplexed intracellular cytokine detection, and measurement signaling pathways through dynamic protein phosphorylation.
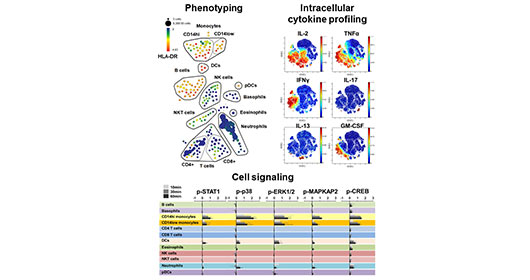
- Flow Cytometry
The HIMC is equipped with a BD LSRFortessa analytical flow cytometer (Becton Dickinson) with high throughput capability (96 well format) to support large volume sample acquisition. We offer a range of flow cytometry assays including myeloma MRD analysis, and identification of all major immune cell types.
- In Vitro Sensitization with Antigen and Expansion of T Lymphocytes
For many antigens, frequency of antigen-specific T cells ex vivo may be too low to reliably measure immune responses. This is true of many tumor antigens but also for some weaker viral antigens. One way to overcome this limitation is to expand antigen specific T cells in vitro using overlapping peptide libraries, preferably independently for CD8 and CD4 T cells that tend to peak at different times after culture. This in vitro sensitization step is designed to only expand in vivo generated T cell responses while minimizing de novo generation of responses from in vitro stimulation, with minimal skewing of repertoires. Although the assay is no longer directly quantitative following presensitization, comparison of eventual frequencies across samples is still possible when assays are performed simultaneously. Besides providing more robust results, presensitization also allows for testing more specificity controls. The expanded cells can be further analyzed using assays such as ELISPOT and intracellular cytokine staining.
- ELISPOT Assay
ELISPOT (Enzyme Linked Immunosorbent SPOT) assay is a robust and standardized test to quantify functional antigen-reactive T cells at the single cell level for capacity to produce cytokines such as IFN-g. PBMCs or purified and/or expanded CD4 or CD8 T cells are stimulated with antigen and cultured in a 96 well plate with a nitrocellulose membrane coated with antibody specific for IFN-g. Results are provided as a number of spots (equivalent to reactive cells) out of cells tested. The test is very sensitive and allows for multiple specificity controls.
- Intracellular staining of cytokines (ICS)
We offer highly multiplexed intracellular cytokine staining by mass cytometry to evaluate the polyfunctionality of antigen specific T cells in conjunction with their marker expression profiles. This approach can be applied directly ex vivo or following presensitization (see above), from PBMC or from purified CD4 and/or CD8 compartments.
- High throughput nucleic acid isolation
We offer high throughput DNA/RNA isolation from clinical specimens using a QiaSymphony system. - High throughput targeted gene expression analysis
The HIMC is equipped with a Fluidigm Biomark HD system and Juno microfluidic controller, which utilize integrated microfluidic circuits to allow high-throughput, low-input qPCR reactions to measure 96 genes on 96 samples. This platform allows high throughput gene expression analysis on cDNA from pooled cells or tissues samples. As one example of the capabilities of the platform, the HIMC offers Fluidigm’s Advanta Immuno-Oncology Gene expression assay, a 170-gene expression qPCR assay targeting key markers of tumor immune response that have been shown to inform tumor progression and checkpoint therapeutic response. - Spatial Transccriptomics
HIMC also provides spatial gene expression analysis using the 10x Visium platform and CytAssist hardware. This next-generation molecular profiling solution allows mapping of the whole transcriptome to distinct spatial structures of a tissue. It allows researchers to gain insight into expression programs and functional states of cells inside preserved tissues at 55 um resolution. Visium is compatible with both archival FFPE and fresh frozen tissues. HIMC has developed and validated experimental and bioinformatics pipelines for efficient profiling of spatially resolved gene expression using Visium fresh frozen and FFPE. - TCRbeta repertoire High Throughput Sequencing with Adaptive Biotechnology
To assess the diversity and clonality of the T cell repertoire, and changes in tumor immunogenicity as a response to treatment, TCRbeta sequencing is performed to correlate with clinical outcomes. The adaptive immune system has the potential to recognize a vast number of antigens through the combinatorial diversity of αβ TCRs. Estimates based on direct sequencing of TCRβ chains indicate that at any one time, an individual carries over 10 million unique TCRβ CDR3 chains. We are developing a platform for in-depth analysis of the T-cell receptor repertoire, a specific and important part of the immune system for the selection, function, and diversity of T cells in collaboration with Adaptive Biotechnology. - High Throughput Single cell sequencing
Conventional ‘bulk’ methods of RNA sequencing (RNA-seq) process hundreds of thousands of cells at a time and average out the differences. However, single cell RNA-seq can reveal the subtle changes that make each cell unique leading to identification of new functionally unique cells. The HIMC offers robust massively-parallel single cell encapsulation using the 10X Genomics Chromium Single Cell platform. This powerful platform allows single cell transcriptomics of hundreds to millions of cells. The platform also enables simultaneous measurement of gene expression together with V(D)J immune repertoires of T and B cells on a cell-by-cell basis.
- MICSSS: Multiplex Immunohistochemical Chromogen Staining on a Single Slide
To address the clinical need for high dimensional analysis of tissues for in situ characterization of immune infiltrates, we had developed a multiplexed chromogen-based IHC staining assay independent of proprietary reagents or equipment that could be readily integrated in routine clinical pathology. MICSSS is performed on FFPE tissue using iterative cycles of staining, revelation, scanning and bleaching of labile chromogenic substrate. It overcomes a major limitation for such high-dimensional analyses, namely tissue availability, by allowing for up to 10 markers to be analyzed from a single tissue slide. The MICSSS method can characterize a large panel of parameters on one single tissue section, including co-localization of markers on single cells while preserving tissue antigenicity and architecture. This tool allow us a comprehensive spatiotemporal map of the local tissue microenvironment, including, i.e.,immunocytes, tumor associated markers, and drivers of immunosuppression. The goal is to offer a comprehensive phenotypic characterization, organization and functionality assessment of samples in relation to clinical parameters and treatment outcome.