Marc and Jennifer Lipschultz Precision Immunology Institute
Microbiome Translational Center
Our Mission at the Microbiome Translation Center (MTC) of the Precision Immunology Institute at the Icahn School of Medicine at Mount Sinai (PrIISM) aims to understand the role of the human microbiome in health and disease and to translate these findings to improve health.
Goals
- Provide centralized research support to incorporate microbiome characterization into planned or existing studies.
- Enhance existing interactions and attract new investigators centered around microbiome-focused studies.
- Translate (pre)-clinical findings to inform medical practice and develop new therapies.
Our Leadership Team
The Microbiome Translational Center (MTC) is comprised of a leadership team with a background in microbiome research, immunology, oncology and computational biology. We work together to offer comprehensive solutions for anything related to microbiome analysis ranging from project development and clinical study/trial design, logistical support in study sample management, a host of cutting-edge microbiome assays and bioinformatics support to assist with the analysis and integration of complex microbiome datasets.
Services
The Microbiome Translation Center’s (MTC) goal is to provide consultation for microbiome studies and serve as a centralize resource hub integrating (pre)-clinical sample collection, biobanking, cryogenic sample processing, nucleotide extraction for microbiome studies and bioinformatic analysis. The MTC also has a state of the (an)-aerobic microbial culturing and mouse gnotobiotics facility.
Services the MTC offers includes:
The MTC can help with your grant application and provide SOPs with regards to sample collection, processing and sequencing as well as computational and statistical data analysis for your microbiome studies.
The MTC is comprised of a leadership team with a background in microbiome research, immunology, oncology and computational biology. We work together to offer comprehensive solutions for anything related to microbiome analysis ranging from project development and clinical study/trial design, logistical support in study sample management, a host of cutting-edge microbiome assays and bioinformatics support to assist with the analysis and integration of complex microbiome datasets.
The MTC facilitates aliquoting and storage of microbiome samples for nucleotide-based analysis and banking of live microbes in collaboration with the existing infrastructure of PrIISM and the Human Immune Monitoring Center (HIMC).
The MTC works with the HIMC for biobanking and sample tracking using a HIPAA compliant secure bar-coded sample management and tracking system using Freezerworks. This system 1) ensures the crucial sample data management in a reliable maintaining sample quality for biomedical research, 2) gets valuable samples to the researchers in a timely and efficient way, and 3) meets all regulatory requirements with an automated audit trail. All the samples are bar-coded with a unique identification number and placed the dedicated storage locations. All storage freezers (-20 and -80oC freezers and LN2 tanks) are installed with temperature monitor “Smart Vue” for the centralized live tracking/remote freezer log-in for 24/7/365 security for the temperature fluctuation. Frozen specimens can be shipped to other institutions using dry-shippers equipped with temperature sensors.
The MTC provides an end-to-end microbiome sample processing service from collection of clinical specimens to a written report.
The MTC services include nucleotide extraction, preparation of sequencing libraries for next-generation sequencing metagenomics and 16S rDNA amplicon sequencing on Illumina platforms. Analysis pipelines focus on existing, open-source and well-validated computational and statistical tools. Results will be presented to investigators as a detailed report that includes description of methods and figures.
- Clinical sample collection. The MTC will provide advice on how best to collect samples from various body sites and prepares SOPs tailored to your study design. Extensive expertise exists for the collection of fecal and oral samples; collection from other body sites are under development.

- Sample processing. Stool samples are aliquoted for microbial culturing and nucleotide extraction under cryogenic conditions using custom equipment ensuring sample stability throughout the process. Nucleotides are extracted using a combination of chemical and mechanical lysis tailored for efficient extraction from a broad range of microorganisms.
- Next-generation sequencing. The MTC provides services for preparation of amplicon 16S rDNA and shotgun metagenomics sequencing libraries for sequencing on Illumina platforms. Additionally, the MTC can generate genome assemblies from pure cultures using Illumina platforms.

- Computational analysis. The Microbiome Translational Center will utilize the high-performance cluster (HPC) resources provided by Scientific Computing at the ISMMS for the analysis of microbiome data. Standard computational pipelines are utilized to process 16S rDNA or shotgun metagenomics data, and results are provided to investigators as a detailed report that includes methods and figures.
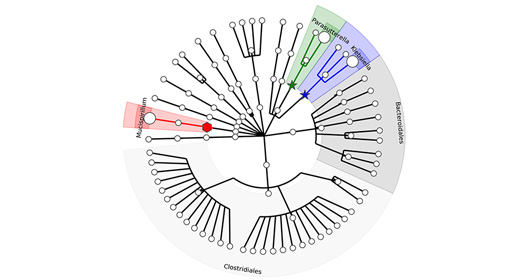

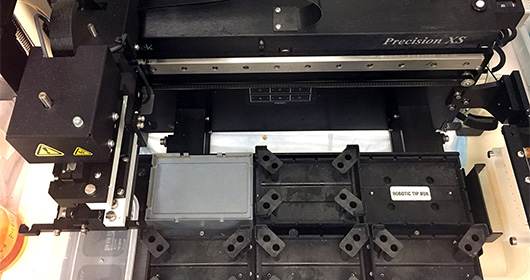
The MTC has a state-of-the-art microbial isolation and cultured facility for anaerobes, microaerophilic and aerobes grown on a variety of conditions that capture 50-80% of the microbes present in a clinical sample. Culturing and isolation of the microbes has a unique advantage over descriptive sequencing techniques in that it allows for genome assembly using SPAde and detailed genome annotation using a variety of tools such as RAST, prokka, CARD/RGI (antibiotic resistance) and BLAST against several virulence databases such as VFanalyzer, Victors, and IslandViewer. More importantly, detailed in vitro and in vivo characterization such as metabolic output of a specific set of microbes and evaluation in various preclinical gnotobiotic disease models. The facility has a double vinyl anaerobic Coy chamber with a Biotek XS robot and utilizes a Bruker MALDI Biotyper for fast species/strain-level validation of the arrayed microbes.

The MTC also offers culturing services (preparation of glycerol stocks, liquid cultures, or gavage-ready suspensions) from clinical and commercially available culture collections e.g. ATCC, DSMZ for use in in vitro or in vivo studies.

Our facility currently accommodates 10 large flexible-film gnotobiotic breeding isolators, each capable of holding 18-24 cages and 16 smaller experimental isolators each capable of housing up to six cages for individual long-term gnotobiotic experiments including those with defined consortia of bacteria, custom diets and specific treatment paradigms. For short-term gnotobiotic experiments the facility has 96 sealed positive pressure gnotobiotic cages as well as the capability for ~100 cages using an outside-the-isolator system for screening gnotobiotic mice with different community compositions. The gnotobiotic facility maintains common strains such as C57BL/6 and Rag2-/- as well as a Swiss-Webster colony for rederivation of new germ-free lines by embryo transfer in collaboration with the Transgenic and Genome Editing CoRE at the Icahn School of Medicine at Mount Sinai. Colonies are tracked in our custom web-based mouse tracking software and database. All germ-free quality control metrics are stored in a web-based digital platform available to the facility operators and maintained in perpetuity.

Instructions
All requests for services are processed through iLabs. Please make sure that you have an iLabs account by registering and logging in.
Publications
Please go here for a full listing of all the publications from the MTC leadership team.
Acknowledgment
The existence of the Microbiome Translation Center depends in part on proper acknowledgment in publications. This is an important metric of the value and this enables us to obtain financial and other support so that we can continue to provide essential services in the best ways possible. It also helps personnel of the Microbiome Translational Center to advance in their careers, adding to the overall health of the Microbiome Translational Center.
We ask investigators that use services of the Microbiome Translational Center to acknowledge this in their publications with the following statement:
- “This works was supported in part through the resources and staff expertise provided by the Microbiome Translational Center at the Icahn School of Medicine at Mount Sinai”.
Co-authorship is the appropriate acknowledgment if contributions from the Microbiome Translational Center involve e.g. major role in study conception and design, development of custom analysis methods, biological interpretation of analyzed results, contribution of intellectual content to manuscript (not only description of methods used) or advanced data analysis that make a publication possible or significantly enhance its value. The Microbiome Translational Center follows accepted scientific criteria for authorship, please refer to the criteria details and for additional criteria information.